
Integration of source databases which use different protein naming conventions is challenging. To ensure maximal connectivity of data we translate each protein name found in the input sources to the most appropriate naming convention available using protein name maps. Naming conventions used throughout the sources form a total order, the strongest one is the primary UniProtKB Swiss-Prot accession (30% of the proteins). If Swiss-Prot name not found we provide UniProtKB TrEMBL accessions (70% of the proteins). The 2018_06 release of UniProtKB was used. To learn more about the UniProtKB database click here.
Naming conventions are ordered and available maps form a tree, as the example image shows below:
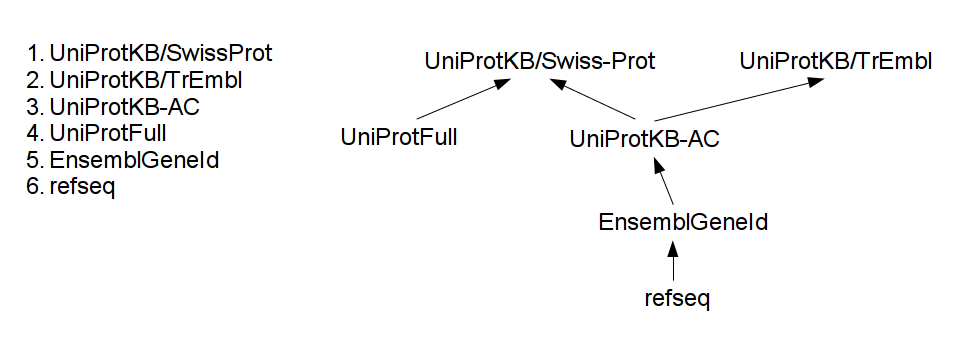
Each time the system encounters a new protein name, it tries to move it as high in the tree as possible, as far the available maps allow it. This method exhaustively travels the mapping information available and provides the best possible integration.
Example: assume a protein-protein interaction source mentions a RefSeq name `NP_005205`. Using the tree of maps, first, it will be translated to its EnsembleGeneId name, `ENSG00000163599`, then comes UniProtKB-AC (which is `P16410`) and at last, the UniProtKB/Swiss-Prot name: P16410. Assume some other protein localization source uses UniProtFull names. Following the same method, `Cytotoxic T-lymphocyte protein 4 precursor` will be translated to P16410, and at the end the two different sources match.
Aside its great performance, this protein name search tree also provides a way to search for synonyms, by traversing the tree top to bottom. ComPPI takes advantage of this and lists several synonyms for each protein.
During this process we used protein ID mapping tables from the UniProtKB and the Human Protein Reference Database and for manual mapping the Synergizer and Protein Identifier Cross-Reference (PICR) webservices.
Please note that, even though the UniProtKB database contains manually annotated Swiss-Prot protein IDs, there are also automatically annotated TrEMBL protein IDs, which could cause redundancy in the data. One protein could be in the database with more than one IDs. Fragments could occur, also in the Swiss-Prot part. Mapping the protein IDs from gene names also could cause bias, such as more than one peptides translated from one gene due to alternative splicing.